Abstract
Expression quantitative trait methylation (eQTM) analysis identifies DNA CpG sites at which methylation is associated with gene expression. The present study describes an eQTM resource of CpG-transcript pairs derived from whole blood DNA methylation and RNA sequencing gene expression data in 2115 Framingham Heart Study participants. We identified 70,047 significant cis CpG-transcript pairs at p < 1E−7 where the top most significant eGenes (i.e., gene transcripts associated with a CpG) were enriched in biological pathways related to cell signaling, and for 1208 clinical traits (enrichment false discovery rate [FDR] ≤ 0.05). We also identified 246,667 significant trans CpG-transcript pairs at p < 1E−14 where the top most significant eGenes were enriched in biological pathways related to activation of the immune response, and for 1191 clinical traits (enrichment FDR ≤ 0.05). Independent and external replication of the top 1000 significant cis and trans CpG-transcript pairs was completed in the Women’s Health Initiative and Jackson Heart Study cohorts. Using significant cis CpG-transcript pairs, we identified significant mediation of the association between CpG sites and cardiometabolic traits through gene expression and identified shared genetic regulation between CpGs and transcripts associated with cardiometabolic traits. In conclusion, we developed a robust and powerful resource of whole blood eQTM CpG-transcript pairs that can help inform future functional studies that seek to understand the molecular basis of disease.
Similar content being viewed by others
Introduction
DNA methylation is an epigenetic modification characterized by the transfer of a methyl group onto DNA cytosine-phosphate-guanine (CpG) sites that can regulate gene expression. The extent of DNA methylation at specific CpG sites is associated with phenotypic variation in numerous traits including cardiovascular disease-related traits1 such as body mass index (BMI)2, blood lipids3, glycemic traits4, blood pressure5, and inflammatory biomarkers6. Expression quantitative trait methylation (eQTM) analysis identifies CpG sites that display methylation-related associations with expression of nearby (cis) or remote (trans) genes. While prior eQTM studies using commercial arrays for gene expression profiling have revealed associations between DNA methylation and gene expression that are linked to clinical disease, many of these studies are limited by small sample size and focus on specific disease phenotypes1,7,8,9.
To generate a resource of eQTM CpG-transcript pairs, we use array-based DNA methylation data and RNA sequencing (RNA-seq) gene expression data in over 2000 Framingham Heart Study (FHS) participants to precisely examine the association between whole blood DNA methylation and gene expression. We posit that leveraging RNA-seq will increase the power, precision, and relevance of our eQTM analyses beyond what was possible with previous array-based gene expression studies7,10. Use of RNA-seq gene expression data also provides an opportunity to interrogate DNA methylation in relation to long non-coding RNAs (lncRNAs) and explore their possible contributions to clinical disease11,12.
The primary aims of this study were to test the associations of DNA methylation with genome-wide gene expression and to create a resource of whole blood eQTM CpG-transcript pairs to facilitate new insights into disease pathways and processes. As proof of principle, we evaluated the associations of eQTM CpG-transcript pairs with cardiovascular traits and provide examples of how these eQTM resources can be used in future research.
Results
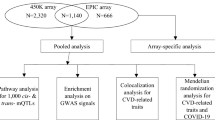
An overview of the study design is presented in Fig. 1. Of 2115 FHS participants (48% women, age 54 ± 15 years) included in the eQTM analysis, 686 were from the FHS Offspring cohort and 1,429 were from the FHS Third Generation cohort (Table 1). The average BMI was 28.0 ± 5.6 kg/m2, while the average serum triglycerides concentration was 114 ± 78 mg/dL. Average fasting blood glucose concentration was 100 ± 21 mg/dL.
Gene-level eQTM results
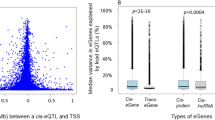
We defined cis CpG-transcript pairs as those where the CpG site and transcription start site were within 1 Mb of one another. We identified 70,047 significant cis CpG-transcript pairs (33,385 unique CpGs; 8534 unique eGenes) at p ≤ 1E−7; the top 10,000 most significant cis CpG transcript pairs are presented in Supplemental Table 1. The majority (49,280; 70%) of the significant cis CpG-transcript pairs involved transcripts of protein-coding genes, while seven percent (4971) were annotated to lncRNAs. The 8534 unique eGenes corresponded to 6693 unique lead CpGs, i.e. the CpG site that is most significantly associated with that eGene in an eQTM CpG-transcript pair. DNA methylation of the lead CpG site was associated with increased cis expression of 2909 (34%) eGenes, and with decreased cis expression of 5625 (66%) eGenes.
We additionally defined trans CpG-transcript pairs as those where the CpG site and transcription start site were > 1 Mb from one another, and we identified 246,667 significant trans CpG-transcript pairs (12,637 unique CpGs, 4300 unique eGenes) at p ≤ 1E−14; the top 10,000 most significant trans CpG-transcript pairs are presented in Supplemental Table 2. The majority (210,944; 86%) of the significant trans CpG-transcript pairs involved transcripts of protein-coding genes, and 11,271 (5%) involved expression of lncRNAs. The 4300 unique eGenes identified corresponded with 1139 lead CpGs. DNA methylation of the lead CpG site was associated with increased trans expression of 1566 (36%) eGenes, and with decreased trans expression of 2734 (64%) eGenes.
A QQ plot comparing observed to expected p-values across all cis and trans CpG-transcript pairs is presented in Supplemental Fig. 1, showing an abundance of significant results and no genomic inflation (inflation λ = 0.998).
Gene-level eQTM internal validation
To conduct internal validation, we divided our total sample into two equal-sized subsets: the first included all participants who had DNA methylation assayed by the 450 K platform (discovery n = 1045), and the second included participants who had DNA methylation assayed by the EPIC platform (validation n = 1070). This internal validation analysis identified 40,148 significant cis CpG-transcript pairs in the discovery 450 K samples at p ≤ 1E−7. Of these, 79% (31,840 CpG-transcript pairs) were significant in the validation EPIC platform at a Bonferroni-corrected p-value ≤ 0.05/40,148. We also identified 241,323 trans CpG-transcript pairs in the discovery sample at p ≤ 1E−14, and of these, 31% (74,405 CpG-transcript pairs) were significant in the validation EPIC platform at a Bonferroni-corrected p-value ≤ 0.05/241,323.
Replication of CpG-transcript pairs
A recent eQTM analysis of nasal epithelial cells by Kim et al.1 identified 16,867 significant CpG-transcript pairs using the Illumina HumanMethylation450K methylation data and RNA-seq gene expression data. We mapped 8481 CpG sites and 3331 gene transcripts to those included in our eQTM analyses, which corresponded to 15,158 CpG-transcript pairs that we subsequently compared with our significant cis and trans eQTM results. Of these, 1192 of 2703 (44%) cis CpG-transcript pairs replicated in our study at p ≤ 1E−7 with matching effect directionality. In contrast, only 41 of 6,165 (0.67%) trans CpG-transcript pairs identified in Kim et al. replicated in our study at p ≤ 1E−14 with matching effect directionality.
For the most significant cis and trans CpG-transcript pairs, we identified the top 1000 most significant unique eGenes and their lead CpG. We evaluated these pairs for replication in two independent external cohorts: a cohort of 1248 participants with European ancestry (EA), African ancestry (AA), and Hispanic ancestry (HA) from the Women’s Health Initiative (WHI), and a cohort of 521 Black participants from the Jackson Heart Study (JHS). Replication results for cis CpG-transcript pairs in both cohorts are presented in Supplemental Table 3, while replication results for trans CpG-transcript pairs in both cohorts are presented in Supplemental Table 4.
In the WHI, all but one of the 1000 cis CpG-transcript pairs from discovery in FHS replicated at a nominally significant p-value of 0.05 with matching effect directionality, while 997 replicated at p < 1E−7 with matching effect directionality. Furthermore, 973 of 1000 trans CpG-transcript pairs from FHS discovery replicated at a nominally significant p-value of 0.05 with matching effect directionality, while 926 trans CpG-transcript pairs replicated at p < 1E−14 with matching effect directionality.
In the JHS, 697 of 820 (85%) cis CpG-transcript pairs from FHS discovery replicated at a nominally significant p-value of 0.05 with matching effect directionality, while 455 (55%) replicated at p < 1E−7 with matching effect directionality. Furthermore, 722 of 880 (82%) trans CpG-transcript pairs from FHS discovery replicated at a nominally significant p-value of 0.05 with matching effect directionality, while 228 (26%) replicated at p < 1E−14 with matching effect directionality.
Gene ontology
We evaluated gene ontology terms to identify specific biological and cellular pathways that were enriched among the top 1000 unique cis and trans eGenes13,14. The top 1000 unique cis eGenes were enriched in 33 pathways related to cell signaling and adhesion at a false discovery rate (FDR) ≤ 0.05 (Supplemental Table 5). The top 1000 unique trans eGenes were enriched in 582 pathways related to the activation of the immune response (Supplemental Table 6).
GWAS enrichment
We examined the overlap of the significant eQTM CpG-transcript pairs with gene-trait associations in the National Human Genome Research Institute (NHGRI)-European Bioinformatics Institute (EBI) GWAS catalog15. Using lead eQTL genetic variants for eQTM transcripts in enrichment analysis, the cis eGenes were enriched for genes associated with 1208 traits in the GWAS catalog at an enrichment FDR ≤ 0.05 (Supplemental Table 7). The trans eGenes were enriched for genes associated with 1191 traits in the GWAS catalog at an enrichment FDR ≤ 0.05 (Supplemental Table 8).
Genomic feature enrichment
We evaluated enrichment of the CpG sites included in significant CpG-transcript pairs for 17 specific genomic features, including CpG islands, shores, bodies, and differentially methylated regions (DMRs). Significant cis CpG-transcript pairs were significantly enriched for 15 of 17 genomic features at an enrichment FDR ≤ 0.05 (Supplemental Table 9), with the top most significant features being upstream CpG shores, CpG islands, downstream CpG shores, DNase hypersensitivity sites (DHS), and gene bodies. Significant trans CpG-transcript pairs were significantly enriched for 13 of 17 genomic features at an enrichment FDR ≤ 0.05 (Supplemental Table 10), with the top most significant features being CpG islands, DHS, gene bodies, regions within 200 bases upstream of the transcription start site (TSS), and enhancers.
eQTM CpG-transcript associations
To illustrate the association between DNA methylation at specific CpG sites and gene expression, we identified the top five most significant cis CpG-transcript pairs and the top five most significant trans CpG-transcript pairs and plotted the associations for each pair (Supplemental Figs. 2 through 11). In general, these figures show a negative association between DNA methylation and gene expression; the only CpG-transcript pair to show a positive association is the trans relationship between methylation at cg13704117 (KIAA1267) and expression of ENSG00000214425.7 (LRRC37A4P) (Supplemental Fig. 11). The CpG-transcript associations in Supplemental Figs. 6, 6, and 8 exhibit bimodal distributions that were not attributable to any nearby SNPs (i.e., within 100 bp) in conditional analysis. rs115687047, located near cg04071440, did not contribute to the bimodality of its cis association with ENSG00000204644.9 (ZFP57) shown in Supplemental Fig. 4 (p = 0.15). rs72989301, located near cg17901463 (GSTM1), was significantly associated with expression of ENSG00000134184.12 (GSTM1) (p = 1.2E−26), but a significant cis association between the CpG and transcript remained (p < 1E−308) as shown in Supplemental Fig. 6. Finally, the SNPs rs1619178 and rs3113782 are located near cg20262684 and have a linkage disequlibrium of 1; they did not contribute to the bimodality of its trans association with ENSG00000185304.15 (RGPD2) shown in Supplemental Fig. 8 (p = 0.75).
Pathways showing cis mediated effects of DNA methylation at cg11024682 on serum triglycerides, fasting blood glucose, and BMI through the expression of SREBF1. The effects of DNA methylation at the CpG site cg11024682 on all three cardiometabolic traits of interest are mediated through cis effects on SREBF1 expression. Coefficients presented in the figure represent the change in gene expression associated with a 10% increase in DNA methylation; the change in serum triglycerides (log(mg/dL)), fasting blood glucose (mmol/L), or BMI (kg/m2) associated with a 10% increase in DNA methylation; or the change in serum triglycerides (log(mg/dL)), fasting blood glucose (mmol/L), or BMI (kg/m2) associated with a 1-unit increase in gene expression.
eQTM CpG-transcript pairs in mediation analysis of cardiometabolic traits
To illustrate the application of the eQTM resource we generated, we provide examples from the associations of significant cis CpG-transcript pairs with three cardiometabolic traits: serum triglycerides, fasting blood glucose, and body mass index (BMI). Specifically, we first evaluated the mediated effect of DNA methylation on the clinical phenotype through the expression of the linked eGene. To identify candidate CpG-transcript pairs to include in mediation analysis, we identified (1) the overlap of significant cis CpG sites with published CpG-trait associations from the MRC Integrative Epidemiology Unit (IEU) EWAS catalog (http://www.ewascatalog.org/)16, and (2) significant results from de novo transcriptome-wide association studies (TWAS) of RNA-seq gene expression data and log-transformed serum triglycerides, fasting blood glucose, and BMI in the FHS. The strategy of aligning trait-associated CpGs with corresponding trait-associated transcripts identified 14 CpG-transcript pairs associated with serum triglycerides, 36 associated with fasting glucose, and 153 associated with BMI. These pairs and their corresponding traits were then tested for the following mediation effects: (1) the total effect of DNA methylation on the cardiometabolic trait, (2) the direct effect of DNA methylation on the cardiometabolic trait independent of gene transcript expression, and (3) the mediated effect of DNA methylation on the cardiometabolic trait through gene transcript expression. Results are summarized in Supplemental Table 11. As an example, Fig. 2 illustrates significant mediation of the effects of DNA methylation at the CpG site cg11024682 on serum triglycerides, fasting blood glucose, and BMI via expression of the SREBF1 gene. The proportion of the association between DNA methylation and the three cardiometabolic traits mediated by SREBF1 expression ranged from 0.16 to 0.36.
Gene transcript expression mediated the effect of DNA methylation on serum triglycerides (log-transformed) for ten of the 14 CpG-transcript pairs. Significant mediation of DNA methylation on gene expression was observed for the expression of RNASET2, SREBF1, ABCG1, and TOM1L2 at p ≤ 0.05.
Gene transcript expression significantly mediated the effect of DNA methylation on fasting blood glucose for 24 of the 36 CpG-transcript pairs, of which six had direct and mediated effects in opposite directions (e.g., a negative direct effect and a positive mediated effect). The greatest proportions of the CpG-fasting glucose associations mediated by gene expression (p < 2E−16 for all) were observed for the expression of KLRF1 (proportion mediated = 0.87), TRAPPC2B (proportion mediated = 0.82), NKIRAS2 (proportion mediated = 0.74), and RRP12 (proportion mediated = 0.52).
Finally, gene transcript expression significantly mediated the effect of DNA methylation on BMI for 117 of the 153 CpG-transcript pairs, of which 55 had direct and mediated effects in opposite directions. For the CpG-transcript pairs where the direct and mediated effects were in the same direction, the greatest proportions of the CpG-BMI associations mediated by gene expression (p < 2E−16 for all) were observed for the expression of TSPYL1 (proportion mediated = 0.98), PLD3 (proportion mediated = 0.94), and FAS (proportion mediated = 0.80). Additionally, two CpG-transcript pairs showed significant mediation of the CpG-BMI association through expression of lncRNA genes BX284668.5 and LINC00996.
eQTM CpG-transcript pairs in colocalization analysis of cardiometabolic traits
We conducted a Bayesian colocalization analysis to evaluate whether the cis-CpG-transcript eQTM pairs associated with serum log triglycerides, fasting blood glucose, and BMI were regulated by shared genetic variants. The MRC IEU EWAS catalog was used to identify CpG sites associated with each of the clinical phenotypes of interest, and three sources of data were used for colocalization analysis (Fig. 3). Among significant serum triglycerides-associated CpG sites identified in the MRC IEU EWAS catalog, 15 overlapped with CpGs from significant cis CpG-transcript pairs where the CpG and gene transcript also shared at least one SNP (i.e., the eQTL variant for the transcript matched the mQTL variant for its paired CpG). Colocalization results for the 15 cis eQTM CpG-transcript pairs, which included 12 unique CpG sites associated with 4876 significant cis-mQTL variants and 11 unique gene transcripts associated with 6,327 significant cis-eQTL variants, are presented in Supplemental Table 12. The probability of colocalization H4 ≥ 80% was observed between the CpG and transcript SNPs for ten eQTM CpG-transcript pairs. The SNP rs7215055 was associated with serum triglycerides17 and was also associated with methylation of CpG cg11024682 and expression of gene ATPAF2 (probability of colocalization H4 = 100%).
Among significant fasting glucose-associated CpG sites identified in the MRC IEU EWAS catalog, 138 overlapped with CpGs from significant cis-CpG-transcript pairs where the CpG and gene transcript also shared at least one SNP. Colocalization results for the 138 cis-eQTM CpG-transcript pairs, which included 109 unique CpG sites associated with 38,758 significant cis-mQTL variants and 172 unique gene transcripts associated with 100,546 significant cis-eQTL variants, are presented in Supplemental Table 13. Probability of colocalization ≥ 80% was observed between the CpG and transcript SNPs for 51 eQTM CpG-transcript pairs. There was no overlap of SNPs identified in a GWAS of fasting glucose18 with both mQTL and eQTL variants for CpG-transcript pairs, thus further colocalization analyses were not carried out for this trait.
Finally, among significant BMI-associated CpG sites identified in the MRC IEU EWAS catalog, 207 overlapped with CpGs from significant cis CpG-transcript pairs where the CpG and gene transcript also shared at least one SNP. Colocalization results for the 207 cis-eQTM CpG-transcript pairs, which included 134 unique CpG sites associated with 82,823 significant cis-mQTL variants and 182 unique gene transcripts associated with 232,511 significant cis-eQTL variants, are presented in Supplemental Table 14. Probability of colocalization ≥ 80% was observed between the CpG and transcript for 67 eQTM CpG-transcript pairs. The SNPs rs3817334, rs10838738, and rs1064608, which were associated with BMI19, were also mQTL genetic variants associated with the CpG cg17580616 and eQTL genetic variants associated with the gene ACP2 (probability of colocalization H4 = 100%).
Discussion
Our study of DNA methylation in relation to gene expression in 2115 FHS participants identified over 70,000 significant cis CpG-transcript pairs and over 246,000 significant trans CpG-transcript pairs. We generated a comprehensive database of whole blood cis and trans eQTM CpG-transcript pairs that can be used to investigate disease mechanisms, pathways, and processes. We previously evaluated eQTM CpG-transcript pairs in the FHS using the Illumina 450 K DNA methylation array in conjunction with array-based gene expression data7. The present study expands on this prior work by incorporating more extensive DNA methylation profiling (Illumina EPIC DNA methylation array) and RNA-seq expression, resulting in a powerful resource for capturing associations between DNA methylation and gene expression. Not only did we identify four times as many cis-eQTM CpG-transcript pairs (70,047 vs. 16,416) and 24% more trans-pairs (246,667 vs. 198,960) than we previously identified using array-based gene expression data7, but we also provide eQTM CpG-transcript pairs for the expression of lncRNAs.
DNA methylation can alter the regulation of gene transcription in multiple ways, such as aiding in the recruitment of proteins involved in increasing gene expression or inhibiting transcription factor binding to a specific DNA sequence. At promoter sites, DNA methylation generally precludes transcription directly by blocking the binding of transcriptional activators or indirectly by recruiting methyl-binding proteins and co-repressor complexes. In this study, we used eQTM analysis to identify numerous CpG sites that, when methylated, are associated with cis and trans eGene expression. Associations between SNPs and DNA methylation (i.e., mQTL associations) may contribute to bimodal distributions in eQTM CpG-transcript associations, many of which were not attributable to any nearby SNPs (i.e., there was no evidence of SNP-on-CpG effects20 contributing to bimodality). Indeed, conditional analyses of nearby SNPs on select associations showed that the CpG-transcript associations are still highly significant. Furthermore, to showcase the potential clinical utility of our findings, we evaluated the role of our eQTM resource when applied to the analysis of three cardiometabolic risk factors—serum triglycerides, fasting blood glucose, and BMI—in mediation and colocalization analyses. We illustrate specific examples of how the eQTM resource can be used to suggest molecular mechanisms linking DNA methylation to clinical phenotypes: Mediation analyses identified putative pathways by which DNA methylation is associated with clinical traits in EWAS, and colocalization analysis suggested shared genetic regulation of CpGs, transcripts, and clinical traits. For example, the results of our mediation analysis showed that expression of the genes ABCG1 and SREBF1 mediates the association between DNA methylation and all three clinical traits.
While the eQTM resource we created is robust and highly replicable, several limitations must be noted. First, FHS is an observational study consisting of participants of predominantly European ancestry; however, we provide evidence of substantial independent external replication of these results, including among individuals with African ancestry from the JHS. Second, while the use of DNA methylation and gene expression data derived from whole blood samples makes our methods more accessible and more easily replicated in other study cohorts, evaluation of CpG-transcript pairs in other tissues may be more relevant to specific disease processes. Additionally, these analyses do not account for linkage disequilibrium and other factors that may correlate proximal CpGs with one another. Finally, this study does not examine the effect of environmental factors on these associations; lifestyle factors such as cigarette smoking are known to affect DNA methylation and gene expression7,21,22,23. Future eQTM research may incorporate environmental data to better understand the epigenetic effects of environmental exposures.
Conclusions
We created a powerful eQTM resource that leverage DNA methylation and RNA-seq data to characterize associations of DNA methylation with gene expression. We provide proof of concept that eQTM resources can be leveraged to explore molecular mechanisms of disease.
Methods
Cohort description
DNA methylation and RNA-seq data were collected from 2115 participants in the FHS Offspring (n = 686) and Third Generation (n = 1429) cohorts. Peripheral whole blood samples were collected in the fasting state at the ninth examination cycle from FHS Offspring participants (2011–2014)24 and at the second examination cycle (2006–2009)25 from FHS Third Generation participants. All study protocols were approved by the Boston Medical Center Institutional Review Board and performed in accordance with the Declaration of Helsinki and relevant regulations, and all study participants provided their written informed consent.
DNA methylation data collection
Buffy coats were isolated from whole blood samples and prepared with bisulfite conversion for the DNA methylation assays. Samples from 1045 FHS Third Generation participants were assayed for DNA methylation using the Infinium HumanMethylation450 BeadChip (Illumina Inc., San Diego, CA). Samples from the remaining 384 FHS Third Generation participants and all FHS Offspring participants were assayed for DNA methylation using the Infinium MethylationEPIC BeadChip (Illumina Inc., San Diego, CA). Methylation probes on autosomal chromosomes were analyzed while probes containing polymorphic SNPs were excluded.
DNA methylation data pre-processing
DNA methylation data were pooled across both the HumanMethylation450 and the MethylationEPIC platforms, which shared 452,568 CpG loci. In addition to the data captured at these loci, samples assayed by the HumanMethylation450 platform had methylation data for 32,945 additional CpG sites, while those assayed by the MethylationEPIC platform had methylation data for 413,524 additional CpG sites.
Quality control (QC) and normalization were performed on the DNA methylation β values using the “dasen” function in the R wateRmelon package (version 1.16.0)26. Normalized β values were then residualized after accounting for batch effects, row effects, column effects, and four PCs constructed from the normalized β values. These residualized β values were subsequently winsorized at the mean ± 3 * standard deviation (SD) of the distribution for each probe.
Ten principal components (PCs) representing technical confounders were identified from the winsorized residuals. These ten PCs and the winsorized β values representing the proportion of methylation per CpG locus were subsequently used in statistical analyses.
RNA-seq data collection
Whole blood samples were collected in PAXgene tubes (PreAnalytiX, Hombrechtikon, Switzerland). Isolation of total RNA was performed according to standard protocols (Asuragen, Inc., Austin, TX) using the PAXgene Blood RNA System Kit as described previously27,28 RNA-seq was performed in accordance with TOPMed protocols (University of Washington Northwest Genomics Center), and the mRNA-seq library was generated using the Illumina TruSeq (Illumina Inc., San Diego, CA). Stranded mRNA kit and sequencing was performed using the Illumina NovaSeq system (Illumina Inc., San Diego, CA). Gene expression was evaluated using RNA SeQC version 2.3.329 and transcript expression was evaluated using RSEM version 1.3.130.
RNA-seq data pre-processing
Gene expression data were normalized using the trimmed mean of M-values (TMM) approach in the R edgeR package31. A log2 transformation was applied to the TMM-normalized values after addition of 1 unit to avoid taking log of zero. This normalized gene expression value was residualized to account for batch effects, RNA concentration, and RNA integrity number.
Statistical methods: eQTM analysis
We identified PCs from the residualized DNA methylation and gene expression data and performed an extensive search of the number of PCs to optimize the cross-validated replication of the CpG-transcript pairs. This study identified ten PCs for the DNA methylation data (percent of variation 53.7%) and five PCs for the gene expression data (percent of variation 51.6%).
We then calculated the association between gene-level CpG sites and gene transcripts to identify significant eQTM CpG-transcript pairs. For each CpG-transcript pair, residualized gene expression was modeled as the outcome with residualized DNA methylation β values as the primary explanatory variable. These models were adjusted for age, sex, white blood cell count, blood cell fraction32, platelet count, five gene expression PCs, and ten DNA methylation PCs. Each CpG-transcript pair association was calculated individually according to this model, which was repeated for each pair using Graphical Processing Units (GPUs) to accelerate computation. Cis gene-level CpG-transcript pairs were defined as those where the CpG site and the eGene (i.e., the gene transcript associated with the CpG) were within 1 Mb kb of one another and were deemed statistically significant at a Bonferroni-corrected p-value of 1E−733. Trans pairs were defined as those where the CpG site and the eGene were more than 1 Mb apart. While an estimated Bonferroni p-value cutoff of 1E−7/50,000 gene transcripts was calculated, we deemed trans pairs as statistically significant at a stricter p-value of 1E−14 to reduce noise and false positives based on an examination of the genomic positions of CpG sites and gene transcripts. This p-value threshold optimized the tradeoff between the replication rate and the number of significant pairs. Results were annotated to the Illumina CpG reference database34 and to GENCODE 30.
Replication of CpG-transcript pairs
In this study, we evaluated replication of the CpG-transcript eQTM pairs identified in two ways. First, we evaluated the overlap of significant cis and trans CpG-transcript pairs with a recent eQTM study that identified 16,867 significant CpG-transcript pairs in nasal epithelial cells (FDR < 0.01). Second, we identified the top 1000 most significant lead CpG sites and their associated eGenes and tested those eQTM pairs for statistical significance in two external and independent study cohorts: the Women’s Health Initiative (WHI) and the Jackson Heart Study (JHS). Both study cohorts completed analyses using DNA methylation data from the Infinium MethylationEPIC Beadchip (Illumina Inc., San Diego, CA) array processed in TOPMed. Background correction and normalization was done using the normal-exponential out-of-band (NOOB) method in the R minfi package35,36. Additionally, both study cohorts used estimated count gene expression data from RNA-SeQC29 from the TOPMed RNA-seq processing pipeline.
Replication cohort: WHI
The WHI is a large prospective study of 161,808 postmenopausal women who enrolled into one of two study components between 1993 and 1998: the clinical trial of hormone therapy, calcium/vitamin D supplementation, and/or dietary modification, and the observational study. The present study included DNA methylation and RNA-seq data from 1248 WHI participants (818 with European ancestry, 338 with African ancestry, and 92 with Hispanic ancestry) who completed the Long Life Study follow-up examination between March 2012 and May 2013. All participants provided their written informed consent. Data cleaning was completed by self-reported race and ethnicity and the eQTM models were adjusted for age at sample collection, race/ethnicity, directly measured CBC counts, 10 expression PCs, and 15 methylation PCs.
Replication cohort: JHS
The JHS is a population-based cohort study of cardiovascular risk factors and disease in 5306 African Americans. The present replication study included DNA methylation and RNA-seq data from 521 JHS participants. All participants provided written informed consent to use of genetic data.
Whole blood samples from the JHS baseline examination were assayed for DNA methylation using the EPIC array as previously described37. The COMBat method38 was used to correct the normalized DNA methylation β values for batch effects.
RNA-seq data from peripheral blood mononuclear cells (PBMCs; average read depth of 50M, University of Washington Northwest Genomics Center (NWGC)) were selected based on expression thresholds of 0.1 transcripts per million (TPM) and 6 expected read counts from RSEM version 1.3.130 in at least 20% of samples. Gene expression values were inverse normal transformed across samples and probabilistic estimation of expression residuals (PEER) was used to generate 10 factors. Gene expression data was subsequently adjusted using age, sex, the top 10 JHS genotype PCs, and the 10 PEER factors.
After QC, 820 of the top 1000 cis CpG-transcript pairs evaluated for replication were available in this cohort; similarly, 880 of the top 1000 trans CpG-transcript pairs evaluated for replication were available in this cohort. eQTM analysis was completed for these pairs that passed QC after adjustment for the fixed effects of age, sex, and CBC, and for family ID as a random effect.
Gene ontology
Entrez gene IDs for the eGenes were included in gene ontology analyses, and we used the function “goana” in the R package limma39 to identify biological, cellular, and molecular pathways enriched in the eGenes. Statistically significant pathways were identified at a false FDR ≤ 0.05.
GWAS enrichment of clinical phenotypes associated with CpG-transcript pairs
The GWAS catalog included associations of 243,587 unique SNPs with 2960 unique clinical traits at p ≤ 5E−8; SNPs were mapped to the gene(s) in which they were located. We compiled a list of unique gene transcripts that were part of statistically significant CpG-transcript pairs, as well as the associations of those genes with clinical phenotypes in the GWAS catalog. We subsequently used a one-sided Fisher’s exact test to evaluate enrichment for each gene with each of the 2960 GWAS traits.
Applications of eQTM resource: mediation analysis
Mediation analysis was performed using the function “mediate” in the R package mediation (version 4.5.0)40 to obtain total, direct, and mediated effects. This analysis provided the proportion of the total effect of the DNA methylation on the clinical phenotype mediated through gene expression. Results were divided by a factor of ten to obtain the effects associated with a 10% increase in DNA methylation of a CpG.
RNA-seq gene expression data for TWAS were residualized after adjustment for technical covariates and pedigree. All TWAS were then adjusted for age, sex, smoking status, alcohol use, white blood cell count, and predicted blood cell fraction32. The TWAS of serum log triglycerides was additionally adjusted for BMI and use of lipid medications, and the TWAS of fasting glucose was additionally adjusted for BMI and diabetes status. Diabetes status was defined as a fasting glucose concentration ≥ 126 mg/dL or use of diabetes medications.
Applications of eQTM resource: colocalization analysis
For the CpG-transcript pairs evaluated in colocalization analysis, we used FHS cis mQTL and cis expression quantitative trait loci (eQTL) results to identify overlapping single nucleotide polymorphisms (SNPs) associated with the CpG and gene transcript, respectively. We then evaluated the overlap of cis-SNPs associated with CpG sites (mQTL) and cis-SNPs associated with gene transcripts (eQTL) with SNPs associated with serum log triglycerides17, fasting blood glucose18, and BMI19 in published genome-wide association studies (GWAS). We used the R package coloc (version 5.1.0)41 to quantify the probabilities (probability of colocalization H4) that a single genetic variant was associated with both DNA methylation and gene expression (i.e., colocalization of the mQTL and eQTL SNPs) and that a single genetic variant was associated with both DNA methylation and the clinical trait of interest (i.e., colocalization of the mQTL and GWAS SNPs).
Data availability
The FHS DNA methylation and RNA-seq datasets analyzed in the current study are available at the database of Genotypes and Phenotypes (dbGaP) repository phs000007.v32.p13 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v30.p11). JHS data are available on dbGaP at phs000286 (phenotype data) and phs000964 (TOPMed data). WHI data are available on dbGaP at phs001237.
References
Kim, S. et al. Expression quantitative trait methylation analysis reveals methylomic associations with gene expression in childhood asthma. Chest 158(5), 1841–1856. https://doi.org/10.1016/j.chest.2020.05.601 (2020).
Samblas, M., Milagro, F. I. & Martínez, A. DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics 14(5), 421–444. https://doi.org/10.1080/15592294.2019.1595297 (2019).
Płatek, T. et al. DNA methylation microarrays identify epigenetically regulated lipid related genes in obese patients with hypercholesterolemia. Mol. Med. (Cambridge, Mass). 26(1), 93. https://doi.org/10.1186/s10020-020-00220-z (2020).
Liu, J. et al. An integrative cross-omics analysis of DNA methylation sites of glucose and insulin homeostasis. Nat. Commun. 10(1), 2581. https://doi.org/10.1038/s41467-019-10487-4 (2019).
Richard, M. A. et al. DNA methylation analysis identifies loci for blood pressure regulation. Am. J. Hum. Genet. 101(6), 888–902. https://doi.org/10.1016/j.ajhg.2017.09.028 (2017).
Myte, R., Sundkvist, A., Van Guelpen, B. & Harlid, S. Circulating levels of inflammatory markers and DNA methylation, an analysis of repeated samples from a population based cohort. Epigenetics 14(7), 649–659. https://doi.org/10.1080/15592294.2019.1603962 (2019).
Yao, C. et al. Epigenome-wide association study of whole blood gene expression in Framingham Heart Study participants provides molecular insight into the potential role of CHRNA5 in cigarette smoking-related lung diseases. Clin. Epigenetics 13(1), 60. https://doi.org/10.1186/s13148-021-01041-5 (2021).
Sharma, N. K. et al. Integrative analysis of glucometabolic traits, adipose tissue DNA methylation, and gene expression identifies epigenetic regulatory mechanisms of insulin resistance and obesity in African Americans. Diabetes 69(12), 2779–2793. https://doi.org/10.2337/db20-0117 (2020).
Maas, S. C. E. et al. Smoking-related changes in DNA methylation and gene expression are associated with cardio-metabolic traits. Clin. Epigenetics 12(1), 157. https://doi.org/10.1186/s13148-020-00951-0 (2020).
Rao, M. S. et al. Comparison of RNA-seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front. Genet. https://doi.org/10.3389/fgene.2018.00636 (2019).
Ismail, N., Abdullah, N., Abdul Murad, N. A., Jamal, R. & Sulaiman, S. A. Long non-coding RNAs (lncRNAs) in cardiovascular disease complication of type 2 diabetes. Diagnostics (Basel, Switzerland). 11(1), 145. https://doi.org/10.3390/diagnostics11010145 (2021).
Maass, P. G., Luft, F. C. & Bähring, S. Long non-coding RNA in health and disease. J. Mol. Med. (Berl.) 92(4), 337–346. https://doi.org/10.1007/s00109-014-1131-8 (2014).
Ashburner, M. et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25(1), 25–29. https://doi.org/10.1038/75556 (2000).
The gene ontology resource: Enriching a GOld mine. Nucleic Acids Res. 49 (D1), D325–D334. https://doi.org/10.1093/nar/gkaa1113 (2021).
Buniello, A. et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47(D1), D1005–D1012. https://doi.org/10.1093/nar/gky1120 (2019).
Battram, T. et al. The EWAS catalog: A database of epigenome-wide association studies. OSF Preprints https://doi.org/10.31219/OSF.IO/837WN (2021).
Richardson, T. G. et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 17(3), e1003062–e1003062. https://doi.org/10.1371/journal.pmed.1003062 (2020).
Chen, J. et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 53(6), 840–860. https://doi.org/10.1038/s41588-021-00852-9 (2021).
Turcot, V. et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 50(1), 26–41. https://doi.org/10.1038/s41588-017-0011-x (2018).
Zhi, D. et al. SNPs located at CpG sites modulate genome-epigenome interaction. Epigenetics 8(8), 802–806. https://doi.org/10.4161/epi.25501 (2013).
Joehanes, R. et al. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 9(5), 436–447. https://doi.org/10.1161/CIRCGENETICS.116.001506 (2016).
Keshawarz, A. et al. Longitudinal change in blood DNA epigenetic signature after smoking cessation. Epigenetics https://doi.org/10.1080/15592294.2021.1985301 (2021).
Huan, T. et al. A whole-blood transcriptome meta-analysis identifies gene expression signatures of cigarette smoking. Hum. Mol. Genet. 25(21), 4611–4623. https://doi.org/10.1093/hmg/ddw288 (2016).
Kannel, W. B., Feinleib, M., McNamara, P. M., Garrison, R. J. & Castelli, W. P. An investigation of coronary heart disease in families: The Framinham offspring study. Am. J. Epidemiol. 110(3), 281–290. https://doi.org/10.1093/oxfordjournals.aje.a112813 (1979).
Tsao, C. W. & Vasan, R. S. Cohort profile: The Framingham Heart Study (FHS): Overview of milestones in cardiovascular epidemiology. Int. J. Epidemiol. 44(6), 1800–1813. https://doi.org/10.1093/ije/dyv337 (2015).
Pidsley, R. et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14(1), 293. https://doi.org/10.1186/1471-2164-14-293 (2013).
Joehanes, R. et al. Gene expression analysis of whole blood, peripheral blood mononuclear cells, and lymphoblastoid cell lines from the Framingham Heart Study. Physiol. Genom. 44(1), 59–75. https://doi.org/10.1152/physiolgenomics.00130.2011 (2011).
Joehanes, R. et al. Gene expression signatures of coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 33(6), 1418–1426. https://doi.org/10.1161/ATVBAHA.112.301169 (2013).
DeLuca, D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics (Oxford, England). 28(11), 1530–1532. https://doi.org/10.1093/bioinformatics/bts196 (2012).
Li, B. & Dewey, C. N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. https://doi.org/10.1186/1471-2105-12-323 (2011).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1), 139–140. https://doi.org/10.1093/bioinformatics/btp616 (2010).
Houseman, E. A. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86. https://doi.org/10.1186/1471-2105-13-86 (2012).
Saffari, A. et al. Estimation of a significance threshold for epigenome-wide association studies. Genet. Epidemiol. 42(1), 20–33. https://doi.org/10.1002/gepi.22086 (2018).
Illumina Inc. Technical Note: Epigenetics—CpG Loci Identification (2010).
Aryee, M. J. et al. Minfi: A flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10), 1363–1369. https://doi.org/10.1093/bioinformatics/btu049 (2014).
Fortin, J. P., Triche, T. J. Jr. & Hansen, K. D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33(4), 558–560. https://doi.org/10.1093/bioinformatics/btw691 (2017).
Raffield, L. M. et al. Coagulation factor VIII: Relationship to cardiovascular disease risk and whole genome sequence and epigenome-wide analysis in African Americans. J. Thromb. Haemost. JTH 18(6), 1335–1347. https://doi.org/10.1111/jth.14741 (2020).
Jiao, C. et al. Positional effects revealed in Illumina methylation array and the impact on analysis. Epigenomics 10(5), 643–659. https://doi.org/10.2217/epi-2017-0105 (2018).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7), e47–e47. https://doi.org/10.1093/nar/gkv007 (2015).
Tingley, D. et al. Mediation: R package for causal mediation analysis. J. Stat. Softw. https://doi.org/10.18637/jss.v059.i05 (2014).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10(5), e1004383–e1004383. https://doi.org/10.1371/journal.pgen.1004383 (2014).
Acknowledgements
This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov). Data on glycemic traits have been contributed by MAGIC investigators and have been downloaded from www.magicinvestigators.org. Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). RNA-seq for “NHLBI TOPMed: Whole Genome Sequencing and Related Phenotypes in the Framingham Heart Study” (phs000974.v1.p1) was performed at the Northwest Genomics Center (HHSN268201600032I). Methylomics for “NHLBI TOPMed: Whole Genome Sequencing and Related Phenotypes in the Framingham Heart Study” (phs000974.v1.p1) was performed at the Keck MGC (HHSN268201600038I). Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity QC, and general program coordination were provided by the TOPMed Administrative Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The Framingham Heart Study (FHS) acknowledges the support of Contracts NO1-HC-25195, HHSN268201500001I and 75N92019D00031 from the National Heart, Lung and Blood Institute and grant supplement R01 HL092577-06S1 for this research. We also acknowledge the dedication of the FHS study participants without whom this research would not be possible. Dr. Vasan is supported in part by the Evans Medical Foundation and the Jay and Louis Coffman Endowment from the Department of Medicine, Boston University School of Medicine. The analytical component of this project was funded by the NHLBI Division of Intramural Research (D. Levy, Principal Investigator). The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts 75N92021D00001, 75N92021D00002, 75N92021D00003, 75N92021D00004, 75N92021D00005. Additional support for TOPMed data was provided through the grant X01 HL139375. We would also like to acknowledge the following: Program Office: (National Heart, Lung, and Blood Institute, Bethesda, Maryland) Jacques Rossouw, Shari Ludlam, Joan McGowan, Leslie Ford, and Nancy Geller Clinical Coordinating Center: (Fred Hutchinson Cancer Research Center, Seattle, WA) Garnet Anderson, Ross Prentice, Andrea LaCroix, and Charles Kooperberg Investigators and Academic Centers: (Brigham and Women's Hospital, Harvard Medical School, Boston, MA) JoAnn E. Manson; (MedStar Health Research Institute/Howard University, Washington, DC) Barbara V. Howard; (Stanford Prevention Research Center, Stanford, CA) Marcia L. Stefanick; (The Ohio State University, Columbus, OH) Rebecca Jackson; (University of Arizona, Tucson/Phoenix, AZ) Cynthia A. Thomson; (University at Buffalo, Buffalo, NY) Jean Wactawski-Wende; (University of Florida, Gainesville/Jacksonville, FL) Marian Limacher; (University of Iowa, Iowa City/Davenport, IA) Jennifer Robinson; (University of Pittsburgh, Pittsburgh, PA) Lewis Kuller; (Wake Forest University School of Medicine, Winston-Salem, NC) Sally Shumaker; (University of Nevada, Reno, NV) Robert Brunner Women’s Health Initiative Memory Study: (Wake Forest University School of Medicine, Winston-Salem, NC) Mark Espeland. LMR and YL are funded by R01AG075884. YL is also funded by R01HL129132. JW was supported by NIH grant 5T32ES007018. The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I) and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute on Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. The funders had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, and in the preparation, review, or approval of the manuscript.
Disclaimer
The views and opinions expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute, the National Institutes of Health, or the U.S. Department of Health and Human Services.
Funding
Open Access funding provided by the National Institutes of Health (NIH).
Author information
Authors and Affiliations
Contributions
R.J., T.H., C.Y., S.H., and D.L. conceived the current study. A.K., R.J., J.M., C.L., C.Y., and D.L. contributed to the design of the study. R.J., C.Y.D., N.L.H., A.N.P., H.L., C.L., J.L., Z.D., M.C., C.P.H., P.C., L.M.R., J.W., Y.L., A.P.R., M.F., N.S., R.S.V., D.L.D., A.P.C., C.K., and D.L. contributed to acquisition of data used in these analyses. A.K., R.J., P.J.M., Y.W., G.M.P., J.H., J.W., and A.P.R. contributed to data analysis. A.K., R.J., C.L., T.H., S.H., B.T., M.S., P.C., P.J.M., and C.Y.D, contributed to data interpretation. A.K. and H.B. drafted the work. J.M., C.L., T.H., B.T., M.S., P.C., P.J.M., C.Y.D., G.M.P., R.S.V., and D.L.D. substantively revised the work. All authors have approved this manuscript.
Corresponding author
Ethics declarations
Competing interests
LMR is a consultant for the TOPMed Administrative Coordinating Center (through Westat). No other authors have competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Keshawarz, A., Bui, H., Joehanes, R. et al. Expression quantitative trait methylation analysis elucidates gene regulatory effects of DNA methylation: the Framingham Heart Study. Sci Rep 13, 12952 (2023). https://doi.org/10.1038/s41598-023-39936-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-39936-3
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.