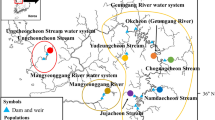
Abstract
Coreius guichenoti is an endemic fish that has been seriously threatened by dam construction in the upper reach of the Yangtze River, and conservation of this species has been a major concern. With a large portion of its spawning grounds disappearing due to dam construction, hatchery release is proposed as a primary strategy for conservation of this species. Genetic information for C. guichenoti is limited, and previous studies are insufficient for robustly detecting genetic structure among reproductively distinct populations. In the present study, the genetic structure of C. guichenoti was investigated using 609 base pairs of the cytochrome c oxidase I gene from drifting larvae and juveniles sampled in three different years. Our results detected two substantial genetic populations of C. guichenoti, with low levels of diversity and strong unidirectional gene-flow between the genetic populations. Based on the results of this study, releasing of juveniles from artificial reproduction should respect genetic differentiation between populations in C. guichenoti: inter-population captive breeding and translocations between the populations should be avoided. Our results suggest that it is crucial to reveal population genetic structures when considering conservation of endemic fish species.




Similar content being viewed by others
References
Ardura A, Gomes V, Linde AR, Moreira JC, Horreo JL, Garcia-Vazquez E (2013) The meeting of waters, a possible shelter of evolutionary significant units for Amazonian fish. Conserv Genet 14:1185–1192
Beerli P (2009) How to use migrate or why are Markov Chain Monte Carlo programs difficult to use? In: Bertorelle G, Bruford MW, Hauffe HC, Rizzoli A, Vernesi C (eds) Population genetics for animal conservation, volume 17 of conservation biology. Cambridge University Press, Cambridge, pp 42–79
Brauer CJ, Unmack PJ, Hammer MP, Adams M, Beheregaray LB (2013) Catchment-scale conservation units identified for the threatened Yarra pygmy perch (Nannoperca obscura) in highly modified river systems. PLoS ONE 8(12):e82953. doi:10.1371/journal.pone.0082953
Carvalho DC, Oliveira DAA, Beheregaray LB, Torres RA (2012) Hidden genetic diversity and distinct evolutionarily significant units in an commercially important Neotropical apex predator, the catfish Pseudoplatystoma corruscans. Conserv Genet 13:1671–1675
Chen DQ, Xiong F, Wang K, Chang YH (2009) Status of research on Yangtze fish biology and fisheries. Environ Biol Fish 85:337–357
Chen YY, Yue PQ (1998) Fauna sinia, osteichthyes-cypriniformes. Science Press, Beijing, pp 329–331
Cheng F, Li W, Wu QJ, Murphy BR, Xie SG (2013a) MOTU analysis of ichthyoplankton biodiversity in the upper Yangtze River, China. J Appl Ichthyol 29:872–876
Cheng F, Liu M, Wu QJ, Xie SG (2013b) Genetic structures of Lepturichthys fimbriata population in the Pingshan section of the Yangtze River. Acta Hydrobiol Sin 37:145–149 (In Chinese)
Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J (2013c) Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30:1224–1228
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Corander J, Marttinen P, Sirén J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinf 9:539
Dudgeon D (2011) Asian river fishes in the Anthropocene: threats and conservation challenges in an era of rapid environmental change. J Fish Biol 79:1487–1524
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics Online 1:47–50
Frankham R (2010) Where are we in conservation genetics and where do we need to go? Conserv Genet 11:661–663
Gwak WS, Nakayama K (2011) Genetic variation of hatchery and wild stocks of the pearl oyster Pinctada fucata martensii (Dunker, 1872), assessed by mitochondrial DNA analysis. Aquac Int 19:585–591
Hammouti N, Schmitt T, Seitz A, Kosuch J, Veith M (2010) Combining mitochondrial and nuclear evidences: a refined evolutionary history of Erebia medusa (Lepidoptera: Nymphalidae: Satyrinae) in Central Europe based on the COI gene. J Zool Syst Evol Res 48:115–125
Hebert PDN, Ratnasingham S, Deward JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Philos Trans R Soc B 270(Suppl):S96–S99
Hedrick PW (2005) Genetics of population, 3rd edn. Jones and Bartlett Publishers, Massachusetts
Huey JA, Espinoza T, Hughes JM (2013) Natural and anthropogenic drivers of genetic structure and low genetic variation in the endangered freshwater cod, Maccullochella mariensis. Conserv Genet 14:997–1008
Institute of Hydrobiology (1976) Fishes of the Yangtze River. Science Press, Beijing, pp 76–78
Kimura M (1980) A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kim WJ, Lee CI, Kim HS, Han HS, Jee YJ, Kong HJ, Nama BH, Kim YO, Kim KK, Kim BS, Lee SJ, Hong KE, Yu JN, Yoon M (2012) Population genetic structure and phylogeography of the ascidian, Halocynthia roretzi, along the coasts of Korea and Japan, inferred from mitochondrial DNA sequence analysis. Biochem Syst Ecol 44:128–135
Leis JM, Carson-Ewart BM (2004) The larvae of indo-pacific coastal fishes, 2nd edn. Brill Press, Boston
Librado P, Rozas J (2009) DnaSP v. 5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu GX, Zhou J, Zhou DG (2012) Mitochondrial DNA reveals low population differentiation in elongate loach, Leptobotia elongata (Bleeker): implications for conservation. Environ Biol Fish 93:393–402
Liu H, Chen D, Liu S, Duan X (2009) Genetic diversity of Botia superciliaris in the upper Yangtze River. Freshw Fish 39:8–13 (in Chinese with English abstract)
Liu LH, Wu GX, Wang ZL (1990) Reproduction ecology of Coreius heterodon (Bleeker) and Coreius guichenoti (Sauvage et Dabry) in the mainstream of the Changjiang River after the construction of Gezgouba dam. Acta Hydrobiol Sin 14:205–215 (In Chinese with English abstract)
Morita T (1999) Molecular phylogenetic relationships of the deep-sea fish genus Coryphaenoides (Gadiformes: Macrouridae) based on mitochondrial DNA. Mol Phylogenet Evol 13:447–454
Park YS, Chang JB, Lek S, Cao WX, Brosse S (2003) Conservation strategies for endemic fish species threatened by the three gorges dam. Conserv Biol 17:1748–1758
Robinson JD, Simmons JW, Williams AS, Moyer GR (2013) Population structure and genetic diversity in the endangered bluemask darter (Etheostoma akatulo). Conserv Genet 14:79–92
Rock J, Costa FO, Walker DI, North AW, Hutchinson WF, Carvalho GR (2008) DNA barcodes of fish of the Scotia Sea, Antarctica indicate priority groups for taxonomic and systematics focus. Antarct Sci 20:253–262
Ryman N (2006) CHIFISH: a computer program testing for genetic heterogeneity at multiple loci using chi-square and Fisher’s exact test. Mol Ecol Notes 6:285–287
Song Z, Song J, Yue B (2008) Population genetic diversity of Prenant’s schizothoracin, Schizothorax prenanti, inferred from the mitochondrial DNA contral region. Environ Biol Fish 81:247–252
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang HY, Yang Z, Gao SH, Chen JS, Zhang YC, Wan L, Qiao Y (2012) Status of fish resources of early life history stages of Coreius guichenoti in the middle reaches of the Jinsha River. Sichuan J Zool 31:416–421 (In Chinese with English abstract)
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequences alignment aided by analysis tools. Nucleic Acids Res 25:4876–4882
Victor BC, Hanner R, Shivji M, Hyde J, Caldow C (2009) Identification of the larval and juvenile stages of the Cubera Snapper, Lutjanus cyanopterus, using DNA barcoding. Zootaxa 2215:24–36
Waples RS (1990) Temporal changes of allele frequency in Pacific salmon: implications for mixed-stock fishery analysis. Can J Fish Aquat Sci 47:968–976
Ward RD, Zemlak TS, Bronwyn H, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc B 360:1847–1857
Weeks AR, Rooyen A, Mitrovski P, Heinze D, Winnard A, Miller AD (2013) A species in decline: genetic diversity and conservation of the Victorian eastern barred bandicoot, Perameles gunnii. Conserv Genet 14:1243–1254
Willi Y, van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Evol Syst 37:433–478
Xu SY, Zhang Y, Wang DQ, Li ZH, Chen DQ (2007) Genetic diversity in largemouth bronze gudgeon (Coreius guichenoti Sauvage et Dabry) from Yibin section of Yangtze River based on sequence of microsatellite DNA. Freshw Fish 37:76–79 (In Chinese with English abstract)
Yang GS, Weng LD, Li LF (2008) Yangtze conservation and development report 2007. Science Press, Beijing, pp 205–213
Yin ZQ, Zhang SY (2008) Thoughts about releasing and proliferation of fisheries resources in China. Chin Fish 3:9–11 (in Chinese with English abstract)
Yu ZT, Liang ZX, Yi BL (1984) The early development of Coreius heterodon and Coreius guihenoti. Acta Hydrobiol Sin 8:370–388 (In Chinese with English abstract)
Yuan XP, Yan L, Xu SY, Wang DQ, Zhang Y, Chen DQ (2008) Genetic diversity of bronze gudgeon (Coreius heterodon) and largemouth bronze gudgeon (C. guichenoti) in Yangtze River basin. J Fish Sci Chin 15:378–385 (In Chinese with English abstract)
Zhang FT, Tan DQ (2010) Genetic diversity in population of largemouth bronze gudgeon (Coreius guichenoti Sauvage et Dabry) from Yangtze River determined by microsatellite DNA analysis. Genes Genet Syst 85:351–357
Zhang YC (2009) Impacts of dam construction on reproduction of Coreius guichenoti and Rhinogobio ventralis in the upper reaches of the Yangtze River. Dissertation, Institute of Hydrobiology, Chinese Academy of Sciences
Acknowledgments
This research was financially supported by the National Science Foundation of China (No. 51209202), Science and Technology Research Project of China Three Gorges Corporation (CT-12-08-01), the special funded project of China Postdoctoral Science Foundation (No. 2013T60764). The participation of Brian R. Murphy was supported by the USDA National Institute of Food and Agriculture (Hatch Project 230537) and by the Acorn Alcinda Foundation, Lewes, Delaware, USA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheng, F., Li, W., Klopfer, M. et al. Population genetic structure and its implication for conservation of Coreius guichenoti in the upper Yangtze River. Environ Biol Fish 98, 1999–2007 (2015). https://doi.org/10.1007/s10641-015-0419-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10641-015-0419-z